Biology 104, 2005 Albert Harris
Fly Genes and Hox Genes
Molecular mechanisms of spatial control of cell differentiation and pattern formation:
What has been learned in Drosophila : (which may or may not be basically the same in vertebrates)
I) Spatial determination is accomplished in a series of different molecular signalling steps,
with separate families of genes responsible for each different step (5 families listed below).
II) At each step, spatial control of gene transcription is controlled by the proteins
coded for by the genes of the previous step. (those of the first step are mostly maternal!)
III) Many of the m-RNA transcripts vary in concentration from one area to another in highly regular patterns
IV) Mutations in a given gene generally result in a failure of formation of whatever structure(s)
normally form(s) from the part of the embryo where this gene's m-RNA is most concentrated.
(what should have been neighboring structures develop there instead)
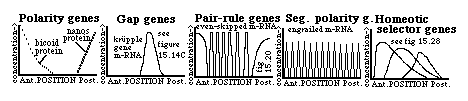
The main families of these (fly) genes are these:
A) Polarity genes (examples include bicoid, nanos, etc.) extreme mutants have tails at both ends
B) Gap genes (krupple) extreme mutants have both sets of end structures, but lack middle structures.
C) Pair-rule genes (even-skipped, fushi terazu) mutants are missing every other segment
D) Segment polarity genes (engrailed) part of every segment is replaced by a mirror image of the rest.
E) Homeotic segment selector genes: mutations cause (certain) imaginal discs to differentiate into structures normally formed only by the imaginal disc of a different segment.
In antennapedia, the antenna disc forms a leg instead; in bithorax, haltere discs form an extra pair of wings.
bicoid RNAs are transcribed by maternal nurse cells, and nanos RNAs by ovarian follicle cells, and transferred from to the extreme anterior end (bicoid) or extreme posterior end (nanos) of the oocyte!
These are therefore said to be maternal effect genes. Translation of these stored RNAs begins at fertilization, and the proteins form diffusion gradients (as drawn above) in the uncleaved early embryo.
The bicoid and nanos proteins are transcription factors, and can bind selectively to the promoter regions of certain other genes, stimulating the transcription of some and inhibiting the transcription of others.
They can also affect translation of certain messengers (for example, Translation of the hunchback gene m-RNA is selectively inhibited by the nanos protein). Combinations of stimulations and inhibitions of transcription cause the spatial distributions of the many different gap genes.;
Likewise, the krupple protein (like other gap gene proteins) is itself a transcription factor: it binds selectively to sites in the promoter regions of still other genes, thus turning them "on" or "off".
Transcription of the krupple gene is inhibited by the hunchback protein, & also by the tailless protein!
Eventually, each location gradually develops a more and more unique combination of gene products.
;
Imagine you are a cell somewhere inside a developing fly embryo, trying to decide which cell type to differentiate into! How to do it? Simply measure the concentrations of all these different gene products at your location. That's the idea. An analogy would be if you were lost in a city where there were dozens of chemical factories, paper mills, perfume manufacturers, cedar wood sawmills, etc. The combinations of smells at each location could tell you exactly where you were located.;
The role of your nose is played by the binding specificities for transcription factors of the DNA base sequences in the promoter sequences for the structural genes of the various luxury proteins. Got it?
There are two sets of Pair-rule genes: a primary set (including "even-skipped") and a secondary set (including fushi terazu). The messengers of both sets appear in 7 stripes! Normal embryos develop 14 segments, but mutants of either class of pair-rule genes fail to develop every other segment. For example, embryos mutated in the fushi terazu gene, form only segments #2,#4,#6,#8,#10,#12 and #14. Guess which segments fail to form as a result of mutations in the gene called "even-skipped"!
Genes with bases sequences too similar to be a coincidence have recently been found in zebra danios, and shown to be expressed in every other pair of somites!!!!!! (reported by one of our job candidates)
Segment polarity genes: such as engrailed, mutations of which cause part of every segment to be replaced by a mirror image of the remainder of the segment. Transcription of the engrailed gene itself is stimulated by the proteins of both the primary and the secondary pair rule genes - hence the 14 bands!
The engrailed gene product seems to help create or maintain the boundaries between parasegments.
Homeotic selector genes: Mutations cause certain imaginal discs differentiate to form what should have been formed by the imaginal disc of a different segment. For example in antennapedia mutants, the antenna disc forms a leg instead. There are two sets of these: those of the antennapedia complex and those of the bithorax complex.
The homeobox (a stereotyped subclass of helix-loop-helix transcription factor base sequence) was first discovered by cloning and sequencing several of these homeotic genes. The "homeobox" is a sequence of 180 bases in some part of the DNA of the gene, while the "homeodomain" is the corresponding 60 amino acid part of the protein (that binds to the DNA of the promoter sequence of the gene whose transcription is being controlled). Note that bicoid, fushi terazu and many other non-homeotic genes also contained homeobox sequences, so these sequences aren't just in genes controlling segmentation.
Flies have one set of homeobox selector genes, arrange in sequence on a chromosome, one after another, with the relative positions of these genes along the chromosome matching the relative positions along the animals' anterior-posterior axes of the morphological effects produced by mutating the genes.
This topological parallel between the relative positions of the genes and the relative positions of their effects is called "co-linearity", and its underlying mechanism/significance has yet to be discovered!
Mammals and frogs turn out to have 4 different sets of hox genes, analogous to the flies' one set.
These are on different chromosomes, with 13 (or a few less genes in each set), but with so much similarity in their base sequences that most can be matched with particular members of the fly series!
The 4 sets are (now) called A, B, C, D; with genes A-1, A-2, A-3 etc.
In situ hybridization maps show these genes are expressed in stripe-like patterns along the body axis of the developing embryo, with the anterior boundaries of each gene's expression obeying co-linearity!
Knock-out experiments (selective elimination of a certain hox gene in transgenic mice) have produced animals that are abnormal in the ways predicted (i.e. loss of structures normally formed just behind the anterior border of expression of the knocked-out gene, with these being replaced by a duplicate set of whatever structures normally form in the segment just posterior to that segment.
Topics for class discussion:
-
1) Is that what you would have expected the effect of a hox gene knock out to be?
2) What mechanisms of control of transcription would require or produce co-linearity?
3) Would you have expected so much conservatism in the base sequences of control genes?
4) Did you see the movie, "The Fly"?
5) Can you suggest how hox genes could control the positions of arms? or fingers?
6) Suggest how such genes could control the shapes of arms and fingers.
7) If a structure can regenerate when cut off, would hox gene reactivation be needed? sufficient?
a) * Nurse cells: mitotic sibling cells of oocytes: 4 incomplete divisions-> 16 cells; cytoplasmic connections One of the 16 becomes the oocyte; the other 15 become nurse cells; most of their cytoplasm goes to oocyte
b) * Shape of the oocyte, is not spherical, but elongated, "sausage shaped". Oocyte axes already determined: anterior-posterior axis and dorso-ventral axis are already decided in the oocyte, with certain maternal effect gene products already being positioned, in contrast to vertebrates, where axes are determined by sperm entry in amphibians, by gravity in birds, and are likewise decided during development in all vertebrates.
c) * Cleavage does not occur for the first 12 mitotic divisions: embryo thus remains syncytial, up until a simultaneous superficial cleavage at the 13th mitosis, forming an approximately 6,000-cell blastoderm.
Until this time m-RNAs and proteins can (sometimes) diffuse through the cytoplasm, forming gradients.
d) The future primordial germ cells form ("pole cells") by an earlier and different type of cleavage, at the posterior end; a special granular kind of cytoplasm becomes localized here, and determines their formation.
e) Gastrulation: endoderm invaginates inward from both ends, meet in middle (sort of as if you formed a stomodeum and a proctodeum, which met each other instead of an archenteron)
Mesoderm is formed by invagination of a tube of cells along the mid-ventral line (very similar to vertebrate neurulation, except that the cells of the tube disperse and do not remain epithelial)
Nerve cells also internalized along the mid-ventral line, but by ingression of two parallel ant-post rows of cells