April 20, 2007
Embryology Biology 441 Spring 2007 Albert Harris
Genetic screens of embryonic lethal genes in flies
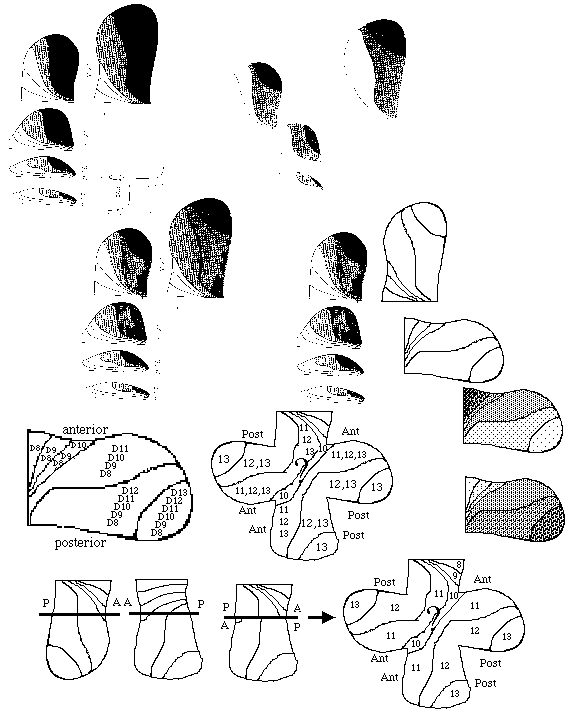
Hox gene expression patterns
A "Genetic Screen" is a systematic search
for mutations that alter some particular property.Directly looking at thousands or millions of individual animals is not an efficient use of time, and cannot find rare mutations.
Researchers therefore invent fiendishly clever "screens" by which properties caused by mutations of interest cause some kind of self-selection.
For example if all individuals that don't have whatever kind of mutation you are looking for will fly away, or die, or disappear, or sink to the bottom, etc.
An analogy would be finding a needle in a hay-stack by using a huge fan to blow away all the hay, or using a giant magnet to pull out anything made of iron.
&&&&&&&&&&&&&&&&&&&&&&&&&&&&&
Long before some interesting gene was mapped, & sequenced, &/or cloned, &/or "knocked-out", (not to mention after being given some cutesy name) some scientist had to discover that gene.
You might think that genome sequencing is how genes get discovered, but that is seldom practical, even in principle.
Nearly always, genes get discovered by isolating mutations in that gene (i.e. organisms in which that gene is mutated).
Only after getting organisms with mutations in a certain gene does it become practical to study the functions and other properties of that gene, such as what it does when NOT mutated.
Probably, you already understand these principles, but lots of people don't seem to get these points (especially in the general public).
The Molecular Biology of Early Fly (Drosophila ) Development:
Yes, I know; flies aren't vertebrates; but historically, much of our modern understanding of genetics, including such important phenomena as genetic linkage, sex linked genes, mutagenesis by X-rays, etc. were first discovered in the fruit fly Drosophila and then later applied to other kinds of animals.
Advantages:
-
short generation times,
smaller genome,
past accumulation of mapping data,
lack of cuteness of the organism (no animal rights complaints!)
make it practical to do research that would be impractical in any vertebrate, especially not in mammals.
The hope is that when we understand how development works in flies, it will turn out to be enough in common with human development to be useful.
1) Fly embryos remain syncytial up to the 6000-plus cell stage, so that proteins, RNA etc. diffuse to create intracellular diffusion gradients.
2) All 3 axes of polarity are already determined during oogenesis:
(anterior-posterior / dorso-ventral / left-right)
[for example, in frog eggs, only one axis is determined in oogenesis;
a second is determined by sperm entry;
and in mammals none of the axes are determined
until much later in development.]
3) Several important mRNAs are synthesized in nurse cells, and some others are synthesized by ovarian follicle cells, and are then positioned by them in certain parts of the fly egg before development begins! (Nothing like this occurs in any vertebrate that I know of!)
4) Flies undergo a metamorphosis from a wing-less worm-like larva into the adult form having wings, legs, antennae, eyes etc. with all of these adult surface structures developing from imaginal discs, which begin as invaginations into the larval ectoderm and then during the time of pupation these discs evert (like turning a glove inside out) and differentiate into the adult's surface structures.
For example, one pair of imaginal discs differentiate into wings, another pair differentiates into forelegs, another pair differentiates into antennae.
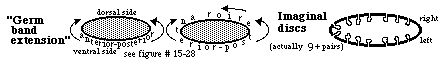
Long ago, a class of homeotic mutations was discovered that causes certain imaginal discs to differentiate into the wrong adult structure: for example the antennapedia mutation caused the antenna imaginal discs to develop into forelegs!
5) Insect gastrulation is rather different from that of other animals you have learned about: the mesoderm is internalized by an infolding along the ventral surface ("the ventral furrow"); the endoderm invaginates from both the posterior and the anterior ends, meeting in the middle!
* * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * *
Other things to keep in mind:
1) Genes are usually named for the defect that results from mutations of the gene!!!!
(For example: if mutating a certain gene caused the heart to fail to develop, then this gene would quite likely be named the heartlessness gene, or "tin man")
The function of the normal version of the gene is to help form the heart; therefore mutations cause them to be unable to perform their normal function. But the gene gets discovered by making mutations in it, and gets named after the effect of the mutation, not the effect of the normal (= "wild type" version of the gene). Remember learning about genes for color-blindness on the human X chromosome? What is the real function of these genes? Is their function to prevent you from seeing colors??
2) Modern molecular biology has techniques for determining the locations of gene products within developing tissues: RNAs are located by "in situ" hybridization with nucleic acids having complementary base sequences; proteins are located by fluorescent (or otherwise labeled) antibodies, often monoclonal antibodies.
Some Generalizations: about the molecular genetics of Drosophila development:
-
a) Spatial determination is accomplished in a series of different steps,
with special families of genes responsible for each step.
b) At each step, spatial control of gene transcription is controlled by the proteins of the genes of the previous step.
c) The m-RNA transcripts vary in concentration from one part of the embryo to another in regular spatial patterns, with the patterns formed at one step controlling those formed at the next step.
d) Mutant forms of a given gene generally result in a failure of formation of whatever structure(s) normally form(s) from the part of the embryo where this gene's m-RNA is most concentrated (neighboring structures spread into the areas where the missing structures should have been)
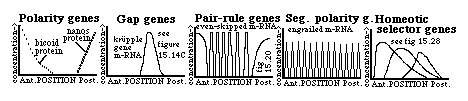
A) Polarity genes (examples include Bicoid, Nanos, etc.) extreme mutants are double-ended
(All these genes are maternal effect genes, since the RNAs are transcribed by maternal nurse cells (bicoid) or ovarian follicle cells (nanos) and transferred from them to either the extreme anterior or the extreme posterior ends of the oocyte!
B) Gap genes (for example, krüpple) extreme mutants have both sets of end structures, but lack certain middle structures.
Transcription of the hunchback gene m-RNA is selectively promoted by the bicoid protein, but translation of this messenger is selectively inhibited by the nanos protein. This controls the eventual distribution of the hunchback protein. On the other hand, transcription of the krupple gene is inhibited by the hunchback protein, and also by the tailless protein (at the rear)!
(Don't bother to memorize these particular genes
Eventually, each location achieves a unique combination of gene products: in other words, if you give me a list of which genes products have high concentrations at your location, and which ones have low concentrations, then I can tell you exactly where you are located, and thus eventually what cell type you should differentiate into, and what tissues and organs you should form there! ( combinatorial rather than cartesian! )
C) Pair-rule genes : two sets of them, in fact, a primary set (including "even-skipped") and a secondary set (including fushi terazu). The messengers of both sets appear in 7 stripes !
Normal embryos develop 14 segments, but mutants of either class of pair-rule genes fail to develop every other segment . For example, embryos mutated in the fushi terazu gene, form only segments #2,#4,#6,#8,#10,#12 and #14. Guess which segments fail to form as a result of mutations in the gene called "even-skipped"!
D) Segment polarity genes: Mutations of these genes (such as engrailed) can cause part of every segment is replaced by a mirror image of the remainder of the segment .
Transcription of the engrailed gene itself is stimulated by the proteins of both the primary and the secondary pair rule genes - hence the 14 bands! The function of the engrailed gene product seems to involve maintenance of the boundaries between parasegments.
E) Homeotic selector genes: Mutations cause certain imaginal discs differentiate to form what should have been formed by the imaginal disc of a different segment .
For example in antennapedia mutants, the antenna disc forms a leg instead. There are two sets of these: those of the antennapedia complex and those of the bithorax complex .
The homeobox (a stereotyped subclass of helix-loop-helix transcription factor base sequence) was first discovered by cloning and sequencing several of these homeotic genes.
The " homeobox " is a sequence of 180 bases in some part of the DNA of the gene, while the "homeodomain" is the corresponding 60 amino acid part of the protein (that binds to the DNA of the promoter sequence of the gene whose transcription is being controlled ). Later, it turned out that bicoid, fushi terazu and many other non-homeotic genes also contained homeobox sequences, so these sequences aren't just in genes controlling segmentation . Many homeotic selector genes contain enormous introns, so big that the time required to transcribe them may possible serve as a timing mechanism!
In the years since these discoveries were made in flies, genes have been discovered in vertebrates and (all?) other phyla, which have DNA base sequences VERY similar to genes in all these families of fly genes.
What does that imply about the most fundamental molecular mechanisms of vertebrate development?
Can these mechanisms be the same as in flies?
Our embryos are never syncytial!
Our development is very regulative, not mosaic!