IUPAC motifs
Twine can use consensus binding sites to identify matches, listed as
one or more strings with the following letters:
A,C,G,T,
R (A or G)
Y (C or T)
S (C or G)
W (A or T)
K (G or T)
M (A or C)
B (C, G, or T)
D (A, G, or T)
H (A, C, or T)
V (A, C, or G)
N (A, C, G, or T).
Initially, the number of mismatches allowed, and the color can be
specified.
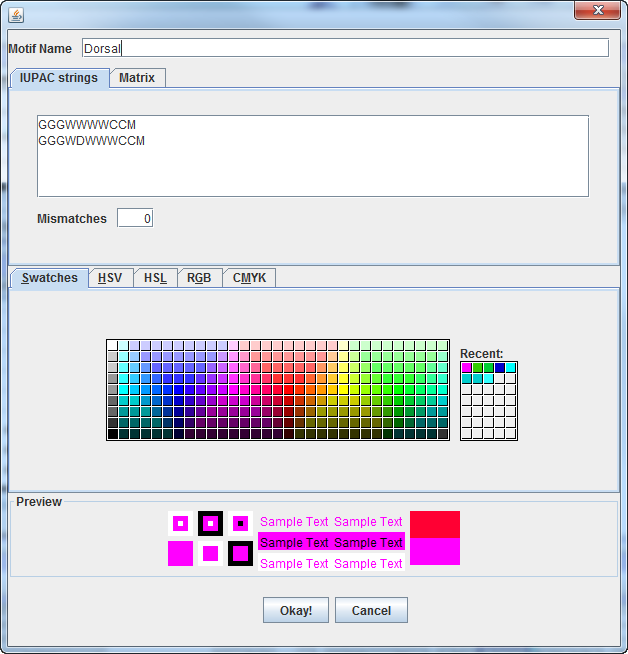
After the motif has been entered, settings to adjust the stringency
can be adjusted in the settings panel:
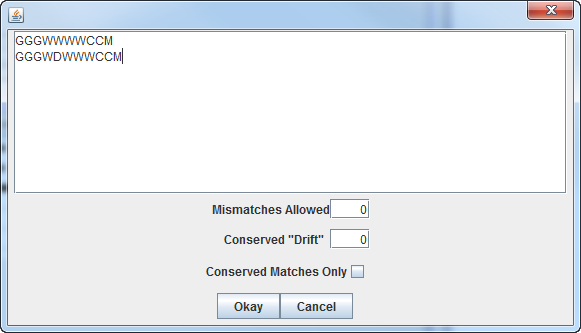
You can add or remove consensus sequences here, and increase the
number of mismatches allowed.
Because alignments are often not perfect, matches in orthologs may
not be aligned to the reference sequence,
so these matches wouldn't be identified as "conserved." Increasing
the "Drift" will allow species matches within
the specified "window" to be considered as part of a linearly
conserved segment.
Selecting "Conserved Matches Only" will filter all matches so that
only the set of matches that pass conservation criteria
(conservation threshold and "drift") will be displayed.

