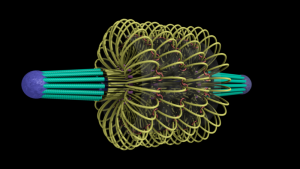
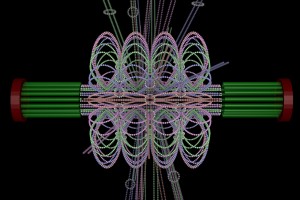
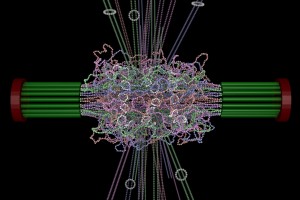
ChromoShake, a three-dimensional simulator designed to find the thermodynamically favored states for given chromosome geometries. The simulator was applied to a geometric model based on experimentally determined characteristics of the budding yeast centromere. In the model shown here, the spindle pole bodies (red disks) and kinetochore microtubules (green rods) depict a metaphase configuration. The colored strands represent the centromere regions of the 16 chromosomes. The centromere chromatin is shown under conditions of thermal fluctuation. Chromosome arms extend perpendicular to the spindle axis. Cohesin (white rings) is radially displaced from the spindle axis. (Image: Josh Lawrimore, Joseph K. Aicher, Patrick Hahn, Alyona Fulp, Ben Kompa, Leandra Vicci, Michael Falvo, Russell M. Taylor, II, and Kerry Bloom, University of North Carolina at Chapel Hill).
 |
 |
End-on view of initial configuration. The spindle microtubules run through the center of the barrel. Cohesin is radially displaced from the spindle axis, its position is determined by the size of the DNA loops.
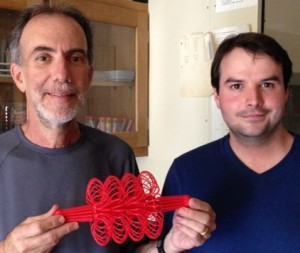
The proud parents Kerry Bloom and Josh Lawrimore hold a 3D printed version of a yeast metaphase spindle printed from a UNC campus facility to make 3D objects of your desire. We are the first to hold a 3D-scale replica of a eukaryotic centromere. Check out additional models and toys that we’ve made.
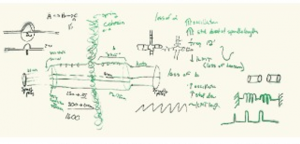
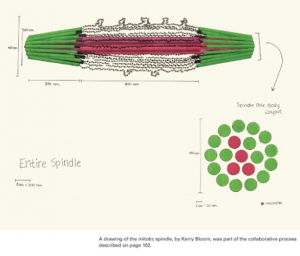
We start with hand drawn models based on experimental evidence. This is a shot of the whiteboard at an early stage in discussion (top). Once we have an image we have our artist render it into a 3D representation of the structure (bottom).
The final stage (above) is to use computer vision to build a geometric model that contains all the elements (microtubules and chromatin) based on their molecular structure and number as experimentally measured. We subject the molecules to thermal motion and ask how well the simulated equilibrium position matches that found in a living cell.
We’ve been known to buy pipe cleaners and cotton balls and spend countless hours making physical models of the centromere (top) before taking the plunge into computer graphics (bottom).